fetal brain MRI motion correction
Papers and Code
Fetpype: An Open-Source Pipeline for Reproducible Fetal Brain MRI Analysis
Dec 19, 2025Fetal brain Magnetic Resonance Imaging (MRI) is crucial for assessing neurodevelopment in utero. However, analyzing this data presents significant challenges due to fetal motion, low signal-to-noise ratio, and the need for complex multi-step processing, including motion correction, super-resolution reconstruction, segmentation, and surface extraction. While various specialized tools exist for individual steps, integrating them into robust, reproducible, and user-friendly workflows that go from raw images to processed volumes is not straightforward. This lack of standardization hinders reproducibility across studies and limits the adoption of advanced analysis techniques for researchers and clinicians. To address these challenges, we introduce Fetpype, an open-source Python library designed to streamline and standardize the preprocessing and analysis of T2-weighted fetal brain MRI data. Fetpype is publicly available on GitHub at https://github.com/fetpype/fetpype.
SUFFICIENT: A scan-specific unsupervised deep learning framework for high-resolution 3D isotropic fetal brain MRI reconstruction
May 26, 2025High-quality 3D fetal brain MRI reconstruction from motion-corrupted 2D slices is crucial for clinical diagnosis. Reliable slice-to-volume registration (SVR)-based motion correction and super-resolution reconstruction (SRR) methods are essential. Deep learning (DL) has demonstrated potential in enhancing SVR and SRR when compared to conventional methods. However, it requires large-scale external training datasets, which are difficult to obtain for clinical fetal MRI. To address this issue, we propose an unsupervised iterative SVR-SRR framework for isotropic HR volume reconstruction. Specifically, SVR is formulated as a function mapping a 2D slice and a 3D target volume to a rigid transformation matrix, which aligns the slice to the underlying location in the target volume. The function is parameterized by a convolutional neural network, which is trained by minimizing the difference between the volume slicing at the predicted position and the input slice. In SRR, a decoding network embedded within a deep image prior framework is incorporated with a comprehensive image degradation model to produce the high-resolution (HR) volume. The deep image prior framework offers a local consistency prior to guide the reconstruction of HR volumes. By performing a forward degradation model, the HR volume is optimized by minimizing loss between predicted slices and the observed slices. Comprehensive experiments conducted on large-magnitude motion-corrupted simulation data and clinical data demonstrate the superior performance of the proposed framework over state-of-the-art fetal brain reconstruction frameworks.
Meta-learning Slice-to-Volume Reconstruction in Fetal Brain MRI using Implicit Neural Representations
May 14, 2025



High-resolution slice-to-volume reconstruction (SVR) from multiple motion-corrupted low-resolution 2D slices constitutes a critical step in image-based diagnostics of moving subjects, such as fetal brain Magnetic Resonance Imaging (MRI). Existing solutions struggle with image artifacts and severe subject motion or require slice pre-alignment to achieve satisfying reconstruction performance. We propose a novel SVR method to enable fast and accurate MRI reconstruction even in cases of severe image and motion corruption. Our approach performs motion correction, outlier handling, and super-resolution reconstruction with all operations being entirely based on implicit neural representations. The model can be initialized with task-specific priors through fully self-supervised meta-learning on either simulated or real-world data. In extensive experiments including over 480 reconstructions of simulated and clinical MRI brain data from different centers, we prove the utility of our method in cases of severe subject motion and image artifacts. Our results demonstrate improvements in reconstruction quality, especially in the presence of severe motion, compared to state-of-the-art methods, and up to 50% reduction in reconstruction time.
HAITCH: A Framework for Distortion and Motion Correction in Fetal Multi-Shell Diffusion-Weighted MRI
Jun 28, 2024



Diffusion magnetic resonance imaging (dMRI) is pivotal for probing the microstructure of the rapidly-developing fetal brain. However, fetal motion during scans and its interaction with magnetic field inhomogeneities result in artifacts and data scattering across spatial and angular domains. The effects of those artifacts are more pronounced in high-angular resolution fetal dMRI, where signal-to-noise ratio is very low. Those effects lead to biased estimates and compromise the consistency and reliability of dMRI analysis. This work presents HAITCH, the first and the only publicly available tool to correct and reconstruct multi-shell high-angular resolution fetal dMRI data. HAITCH offers several technical advances that include a blip-reversed dual-echo acquisition for dynamic distortion correction, advanced motion correction for model-free and robust reconstruction, optimized multi-shell design for enhanced information capture and increased tolerance to motion, and outlier detection for improved reconstruction fidelity. The framework is open-source, flexible, and can be used to process any type of fetal dMRI data including single-echo or single-shell acquisitions, but is most effective when used with multi-shell multi-echo fetal dMRI data that cannot be processed with any of the existing tools. Validation experiments on real fetal dMRI scans demonstrate significant improvements and accurate correction across diverse fetal ages and motion levels. HAITCH successfully removes artifacts and reconstructs high-fidelity fetal dMRI data suitable for advanced diffusion modeling, including fiber orientation distribution function estimation. These advancements pave the way for more reliable analysis of the fetal brain microstructure and tractography under challenging imaging conditions.
SpaER: Learning Spatio-temporal Equivariant Representations for Fetal Brain Motion Tracking
Jul 29, 2024


In this paper, we introduce SpaER, a pioneering method for fetal motion tracking that leverages equivariant filters and self-attention mechanisms to effectively learn spatio-temporal representations. Different from conventional approaches that statically estimate fetal brain motions from pairs of images, our method dynamically tracks the rigid movement patterns of the fetal head across temporal and spatial dimensions. Specifically, we first develop an equivariant neural network that efficiently learns rigid motion sequences through low-dimensional spatial representations of images. Subsequently, we learn spatio-temporal representations by incorporating time encoding and self-attention neural network layers. This approach allows for the capture of long-term dependencies of fetal brain motion and addresses alignment errors due to contrast changes and severe motion artifacts. Our model also provides a geometric deformation estimation that properly addresses image distortions among all time frames. To the best of our knowledge, our approach is the first to learn spatial-temporal representations via deep neural networks for fetal motion tracking without data augmentation. We validated our model using real fetal echo-planar images with simulated and real motions. Our method carries significant potential value in accurately measuring, tracking, and correcting fetal motion in fetal MRI sequences.
A Literature Review on Fetus Brain Motion Correction in MRI
Jan 30, 2024

This paper provides a comprehensive review of the latest advancements in fetal motion correction in MRI. We delve into various contemporary methodologies and technological advancements aimed at overcoming these challenges. It includes traditional 3D fetal MRI correction methods like Slice to Volume Registration (SVR), deep learning-based techniques such as Convolutional Neural Networks (CNNs), Long Short-Term Memory (LSTM) Networks, Transformers, Generative Adversarial Networks (GANs) and most recent advancements of Diffusion Models. The insights derived from this literature review reflect a thorough understanding of both the technical intricacies and practical implications of fetal motion in MRI studies, offering a reasoned perspective on potential solutions and future improvements in this field.
SE-Equivariant and Noise-Invariant 3D Motion Tracking in Medical Images
Dec 21, 2023



Rigid motion tracking is paramount in many medical imaging applications where movements need to be detected, corrected, or accounted for. Modern strategies rely on convolutional neural networks (CNN) and pose this problem as rigid registration. Yet, CNNs do not exploit natural symmetries in this task, as they are equivariant to translations (their outputs shift with their inputs) but not to rotations. Here we propose EquiTrack, the first method that uses recent steerable SE(3)-equivariant CNNs (E-CNN) for motion tracking. While steerable E-CNNs can extract corresponding features across different poses, testing them on noisy medical images reveals that they do not have enough learning capacity to learn noise invariance. Thus, we introduce a hybrid architecture that pairs a denoiser with an E-CNN to decouple the processing of anatomically irrelevant intensity features from the extraction of equivariant spatial features. Rigid transforms are then estimated in closed-form. EquiTrack outperforms state-of-the-art learning and optimisation methods for motion tracking in adult brain MRI and fetal MRI time series. Our code is available at github.com/BBillot/equitrack.
Self-supervised Fetal MRI 3D Reconstruction Based on Radiation Diffusion Generation Model
Oct 16, 2023



Although the use of multiple stacks can handle slice-to-volume motion correction and artifact removal problems, there are still several problems: 1) The slice-to-volume method usually uses slices as input, which cannot solve the problem of uniform intensity distribution and complementarity in regions of different fetal MRI stacks; 2) The integrity of 3D space is not considered, which adversely affects the discrimination and generation of globally consistent information in fetal MRI; 3) Fetal MRI with severe motion artifacts in the real-world cannot achieve high-quality super-resolution reconstruction. To address these issues, we propose a novel fetal brain MRI high-quality volume reconstruction method, called the Radiation Diffusion Generation Model (RDGM). It is a self-supervised generation method, which incorporates the idea of Neural Radiation Field (NeRF) based on the coordinate generation and diffusion model based on super-resolution generation. To solve regional intensity heterogeneity in different directions, we use a pre-trained transformer model for slice registration, and then, a new regionally Consistent Implicit Neural Representation (CINR) network sub-module is proposed. CINR can generate the initial volume by combining a coordinate association map of two different coordinate mapping spaces. To enhance volume global consistency and discrimination, we introduce the Volume Diffusion Super-resolution Generation (VDSG) mechanism. The global intensity discriminant generation from volume-to-volume is carried out using the idea of diffusion generation, and CINR becomes the deviation intensity generation network of the volume-to-volume diffusion model. Finally, the experimental results on real-world fetal brain MRI stacks demonstrate the state-of-the-art performance of our method.
AFFIRM: Affinity Fusion-based Framework for Iteratively Random Motion correction of multi-slice fetal brain MRI
May 12, 2022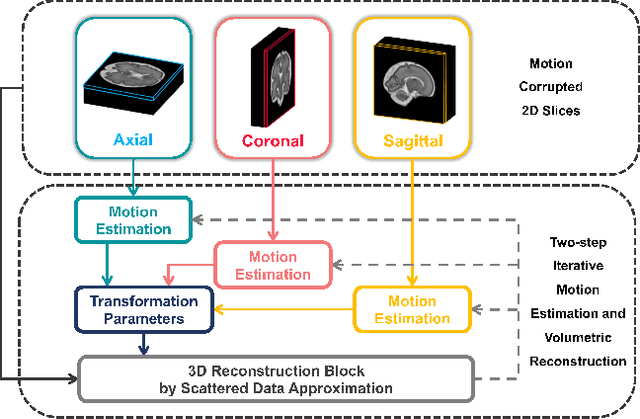
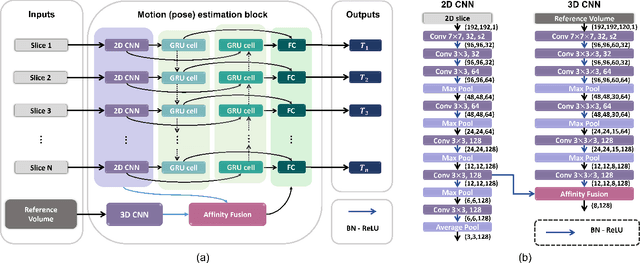
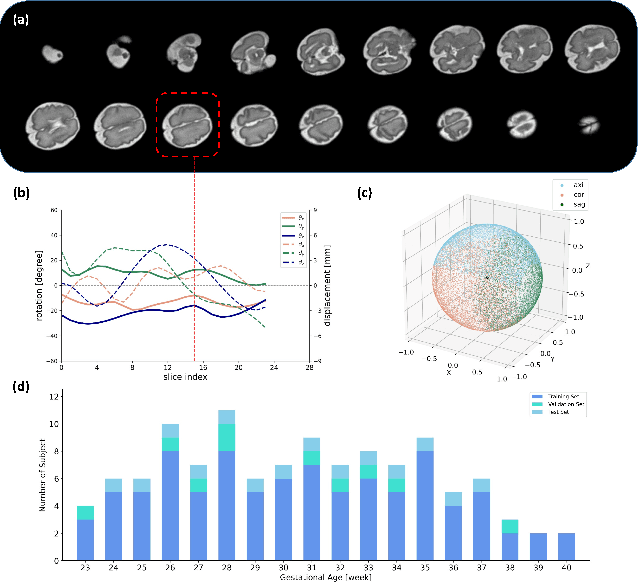
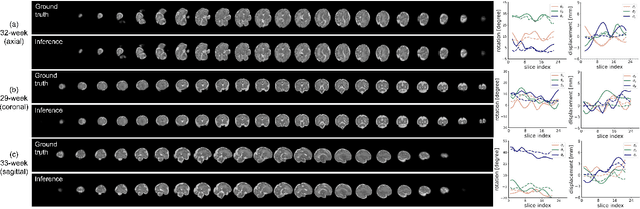
Multi-slice magnetic resonance images of the fetal brain are usually contaminated by severe and arbitrary fetal and maternal motion. Hence, stable and robust motion correction is necessary to reconstruct high-resolution 3D fetal brain volume for clinical diagnosis and quantitative analysis. However, the conventional registration-based correction has a limited capture range and is insufficient for detecting relatively large motions. Here, we present a novel Affinity Fusion-based Framework for Iteratively Random Motion (AFFIRM) correction of the multi-slice fetal brain MRI. It learns the sequential motion from multiple stacks of slices and integrates the features between 2D slices and reconstructed 3D volume using affinity fusion, which resembles the iterations between slice-to-volume registration and volumetric reconstruction in the regular pipeline. The method accurately estimates the motion regardless of brain orientations and outperforms other state-of-the-art learning-based methods on the simulated motion-corrupted data, with a 48.4% reduction of mean absolute error for rotation and 61.3% for displacement. We then incorporated AFFIRM into the multi-resolution slice-to-volume registration and tested it on the real-world fetal MRI scans at different gestation stages. The results indicated that adding AFFIRM to the conventional pipeline improved the success rate of fetal brain super-resolution reconstruction from 77.2% to 91.9%.
Deep Learning Framework for Real-time Fetal Brain Segmentation in MRI
May 02, 2022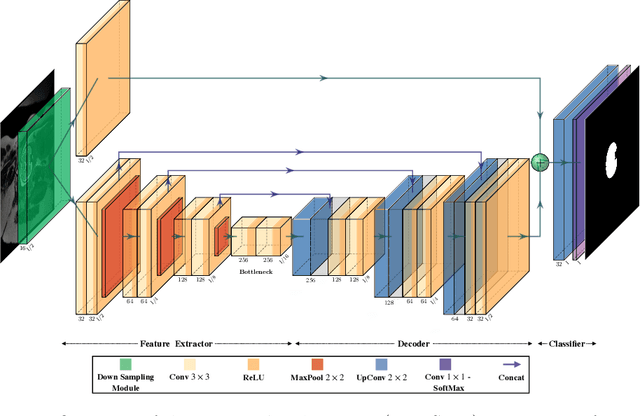
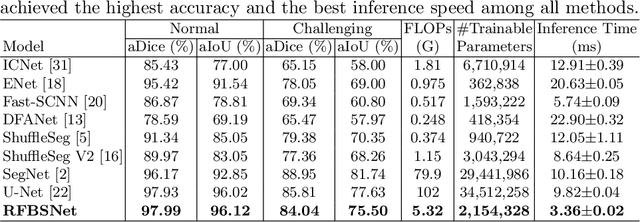
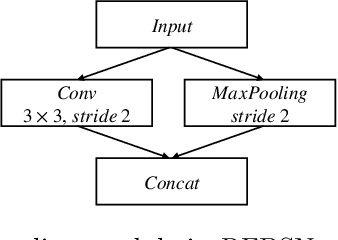
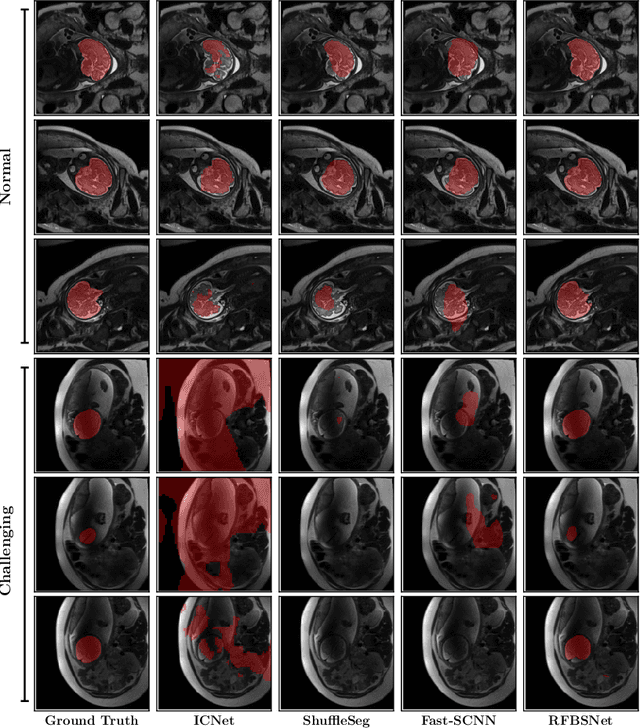
Fetal brain segmentation is an important first step for slice-level motion correction and slice-to-volume reconstruction in fetal MRI. Fast and accurate segmentation of the fetal brain on fetal MRI is required to achieve real-time fetal head pose estimation and motion tracking for slice re-acquisition and steering. To address this critical unmet need, in this work we analyzed the speed-accuracy performance of a variety of deep neural network models, and devised a symbolically small convolutional neural network that combines spatial details at high resolution with context features extracted at lower resolutions. We used multiple branches with skip connections to maintain high accuracy while devising a parallel combination of convolution and pooling operations as an input downsampling module to further reduce inference time. We trained our model as well as eight alternative, state-of-the-art networks with manually-labeled fetal brain MRI slices and tested on two sets of normal and challenging test cases. Experimental results show that our network achieved the highest accuracy and lowest inference time among all of the compared state-of-the-art real-time segmentation methods. We achieved average Dice scores of 97.99\% and 84.04\% on the normal and challenging test sets, respectively, with an inference time of 3.36 milliseconds per image on an NVIDIA GeForce RTX 2080 Ti. Code, data, and the trained models are available at https://github.com/bchimagine/real_time_fetal_brain_segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge